An experiment looking into the effects on mapping statistics and SNV calling with bwa kmer
-ksizes 16-23.
Conclusions
SNV Calling
The effect of altering
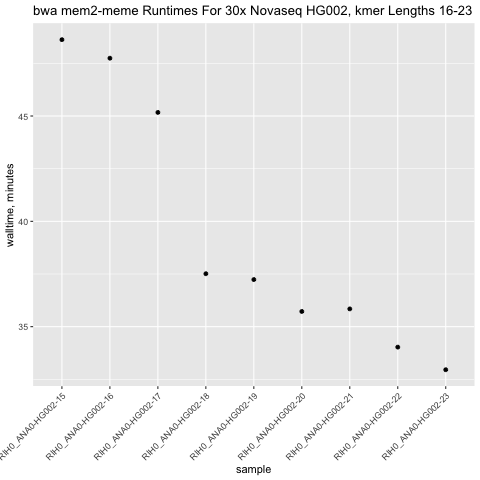
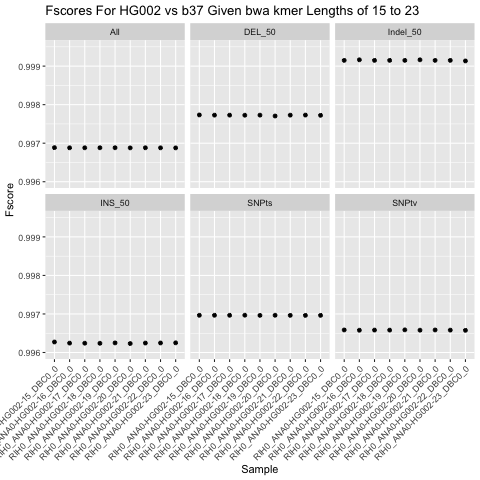
bwa mem2 memekmer-ksizes has neglible effects on calling all classes of SNVs (SNPts, SNPtv, INS, DEL, Indel), with a small gain seen in-k 16. However, this gain comes at a 30% (47m vs 37m) increase in mapping time vs the default-k 19.-kof 15 improves calling vs the default of-k19 by adding 1 TP, calling 16 fewer FP and 1 less FN. This improvement is likely not worth the substantially longer runtime of bwa.
Alignment Metrics
Of the 167 mapping alignment statistics calculated by alignstats, few are significantly different with changing
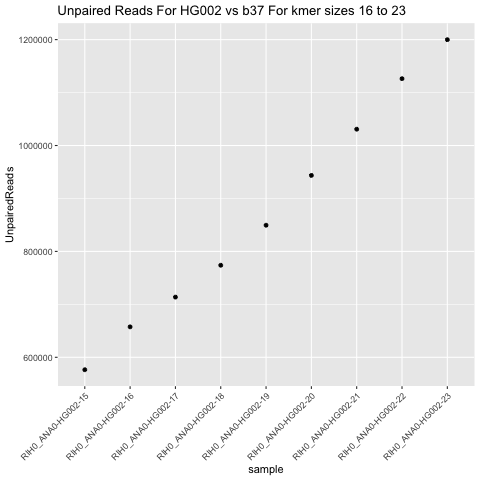
-k. However, a few, such as the number of unpaired reads does decrease as-kbecomes smaller.
SV Calling Impacts?
SV calling has not been assessed here, but the sizable decrease in unpaired reads at
-k 15may indicate SV calling would improve with lower-k.
Methods
- The daylily framework was run on the 30x Novaseq google brain fastqs for HG002.
- bwa mem2 meme was run with
-kvalues from 16 to 23.-k 19being the default. - Deduplication was run with
doppelmarkand variant calling bydeepvariant(see v0.5.3 daylily release for commands). - Alignment statistics calculated with
alignstats.
Results
bwa mem2 meme Runtimes
for -k 16 to 23
Number Unpaired Reads
for -k 16 to 23
SNV Performance w/Deepvariant
for -k 16 to 23
SNV Raw Data
|SAMPLE-bwa kmer|Target Region Size|TN|FN|TP|FP|Fscore| | — | — | — | — | — | — | — | |HG002-15| 2525727871| 2521853400| 19944| 3854527| 4148| 0.9968845797| |HG002-16| 2525727871| 2521853397| 19945| 3854529| 4177| 0.996880714| |HG002-17| 2525727871| 2521853399| 19950| 3854522| 4168| 0.996881224| |HG002-18| 2525727871| 2521853400| 19949| 3854522| 4164| 0.9968818686| |HG002-19| 2525727871| 2521853400| 19945| 3854526| 4164| 0.9968823874| |HG002-20| 2525727871| 2521853397| 19946| 3854528| 4189| 0.9968790374| |HG002-21| 2525727871| 2521853396| 19951| 3854524| 4164| 0.9968816123| |HG002-22| 2525727871| 2521853397| 19948| 3854526| 4179| 0.996880067| |HG002-23| 2525727871| 2521853397| 19948| 3854526| 4189| 0.996878778|